Sensory organs
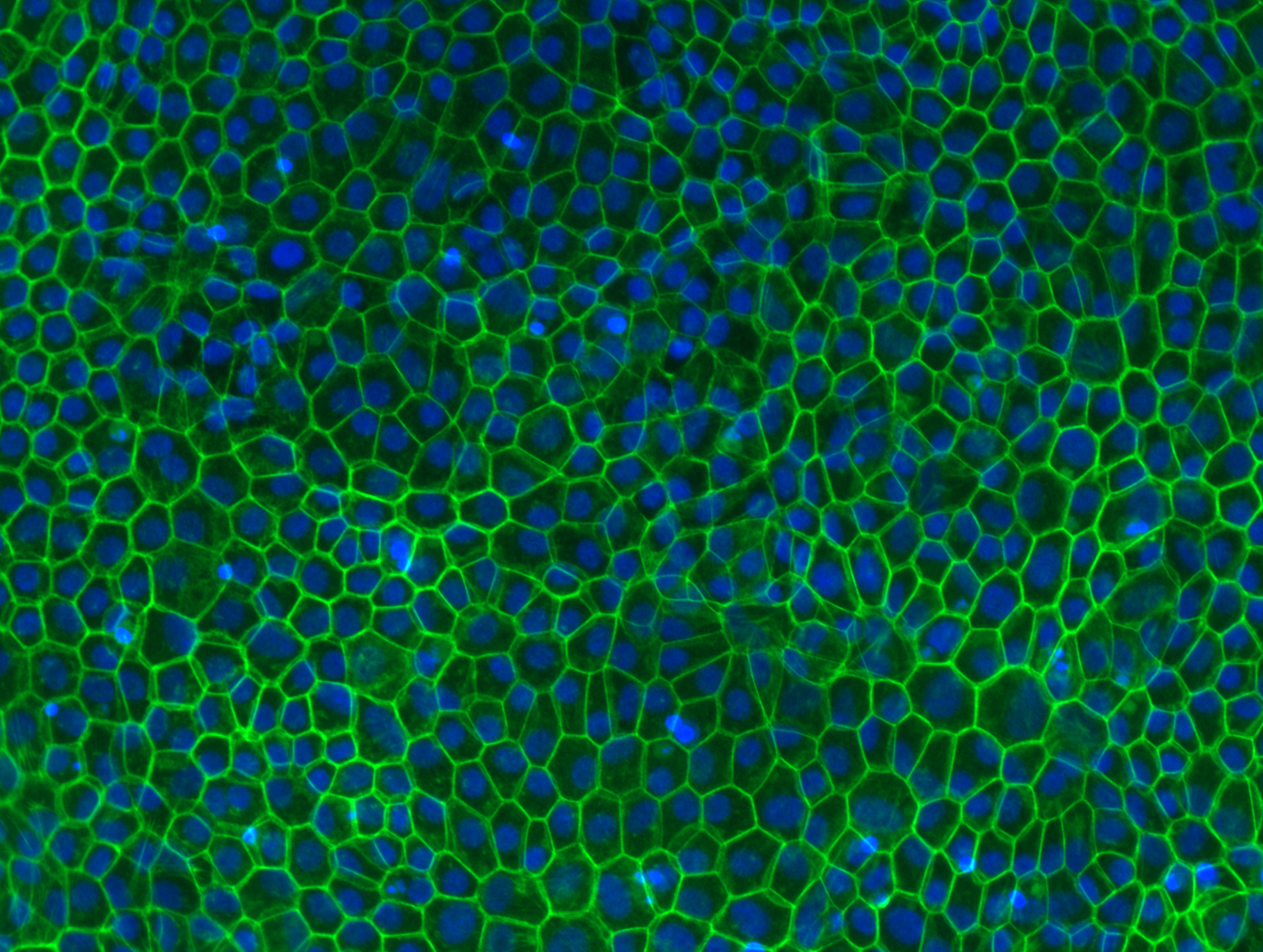
iPSC-derived retinal pigment epithelial cells stained with phalloidin (actin) and DAPI (nuclei).
Courtesy of the Grimm lab
____________________________________________________________
LABS |
|
|
_____________________________________________________________
|
|
|
|
|
Professor for Experimental Ophthalmology, University of Zurich Head of Lab for Retinal Cell Biology, Dept. Ophthalmology, University Hospital Zürich Research Focus: Our research focuses on blinding diseases of the retina with a special focus on age related macular degeneration (AMD) that affects up to 25% of people above the age of 75. We use various animal models to understand specific aspects of disease etiology and progression, and develop therapeutic strategies that are based on neuroprotection and AAV-mediated gene therapy. Photoreceptor neurons form a functional unit with the underlying cells of the retinal pigment epithelium (RPE) that not only form the outer blood/retina barrier but also are crucial for photoreceptor function and survival through the regulation of metabolic support and oxygen delivery from the choroidal blood vessels to photoreceptors. Part of this regulation is the transport and recycling of lipids that originate from the daily process of photoreceptor renewal. We are using iRPE cells differentiated from iPSCs derived from patients carrying genetic variants that confer either increased or decreased risk to develop AMD. These cells closely resemble the in vivo situation in patients and are ideal to study their function in response to various stimuli and conditions that include exposure to neuronal debris with a high cholesterol load and hypoxia, an important factor for disease development. We also use CRISPR editing to inactivate genes in isogenic cells in order to study specific genetic factors for RPE function. Methods: differentiation of iPSC into iRPE cells, CRISPR/Cas9 genome editing, cholesterol efflux, phagocytosis, transepithelial resistance, single cell sequencing, transcriptomics, proteomics, AAV-based gene therapy. Keywords: RPE, lipids, hypoxia, iPSC, neurons, gene therapy Topics: neurodegeneration, gene therapy Publications: http://home.ggaweb.ch/LabForRetinalCellBiology/page1/page1.html Website: http://home.ggaweb.ch/LabForRetinalCellBiology/index.html _________________________________________________________________________ |
