Neuroscience
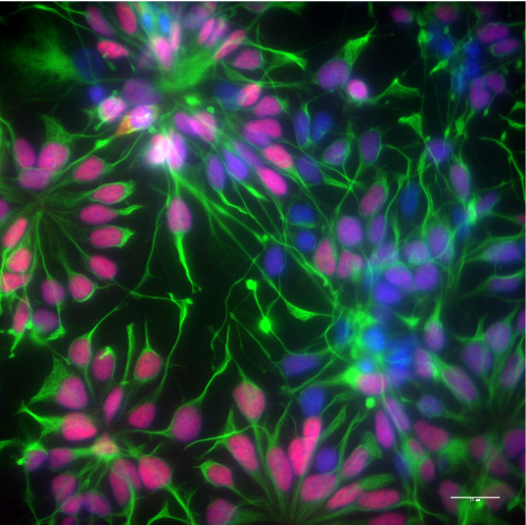
Neural stem cells forming rosettes, stained against the forebrain transcription factor PAX6 (red) and the intermediate neurofilament Nestin (green).
Courtesy of Bachmann lab (Affef Abidi)
__________________________________________________________
LABS
| Ruxandra Bachmann-Gagescu | Christian Grimm | |
| Edna Grünblatt | Sebastian Jessberger | Magdalini Polymenidou |
| Gerhard Schratt | ChristianTackenberg | Janos Vörös |
_______________________________________________________
|
|
Prof.Dr.med. Ruxandra Bachmann-Gagescu Associate Professor for Developmental Genetics Department of Molecular Life Sciences and Institute of Medical Genetics, University of Zurich Research Focus: Our research focuses on a group of human Mendelian disorders called ciliopathies, which are unified by shared genetic causes resulting in primary cilium dysfunction. Primary cilia are small non-motile organelles present on the surface of most vertebrate cells where they are involved in transduction of sensory, mechanical or chemical signals and in regulation of signaling pathways during development and cell homeostasis. Typical clinical presentations of ciliopathies include neurological involvement, retinal degeneration and renal fibrocystic disease, as illustrated by Joubert syndrome (JS), an iconic ciliopathy, which is the main focus of our research. To understand the consequences of mutations in JS-associated genes at the molecular level, we are developing iPSC-based neuronal 2D and 3D models using CRISPR/Cas9 genome editing, imaging and –omics approaches such as RNAsequencing and proteomics. Methods: CRISPR/Cas9 genome editing, next generation sequencing for clone selection and quality control, 2D cortical neuronal differentiation, 3D cerebral organoids Keywords: ciliopathies, primary cilia, Joubert syndrome, iPSCs, neurons Topics: Neuroscience, Disease Modelling Publications: https://pubmed.ncbi.nlm.nih.gov/?term=ruxandra+Bachmann-Gagescu&sort=date&size=100 Website: https://www.medgen.uzh.ch/en/forschung/gagescu.html _____________________________________________________________________top |
|
|
Professor for Experimental Ophthalmology, University of Zurich Head of Lab for Retinal Cell Biology, Dept. Ophthalmology, University Hospital Zürich Research Focus: Our research focuses on blinding diseases of the retina with a special focus on age related macular degeneration (AMD) that affects up to 25% of people above the age of 75. We use various animal models to understand specific aspects of disease etiology and progression, and develop therapeutic strategies that are based on neuroprotection and AAV-mediated gene therapy. Photoreceptor neurons form a functional unit with the underlying cells of the retinal pigment epithelium (RPE) that not only form the outer blood/retina barrier but also are crucial for photoreceptor function and survival through the regulation of metabolic support and oxygen delivery from the choroidal blood vessels to photoreceptors. Part of this regulation is the transport and recycling of lipids that originate from the daily process of photoreceptor renewal. We are using iRPE cells differentiated from iPSCs derived from patients carrying genetic variants that confer either increased or decreased risk to develop AMD. These cells closely resemble the in vivo situation in patients and are ideal to study their function in response to various stimuli and conditions that include exposure to neuronal debris with a high cholesterol load and hypoxia, an important factor for disease development. We also use CRISPR editing to inactivate genes in isogenic cells in order to study specific genetic factors for RPE function. Methods: differentiation of iPSC into iRPE cells, CRISPR/Cas9 genome editing, cholesterol efflux, phagocytosis, transepithelial resistance, single cell sequencing, transcriptomics, proteomics, AAV-based gene therapy. Keywords: RPE, lipids, hypoxia, iPSC, neurons, gene therapy Topics: neurodegeneration, gene therapy Publications: http://home.ggaweb.ch/LabForRetinalCellBiology/page1/page1.html Website: http://home.ggaweb.ch/LabForRetinalCellBiology/index.html _________________________________________________________________________top |
|
|
Head of the Translational Molecular Psychiatry, University Hospital of Psychiatry (PUK), Department of Child and Adolescent Psychiatry and Psychotherapy, University of Zurich Chair of the ECNP Thematic Working Group iPSCs platform for Neuropsychiatry Research Focus: The main research focus of the Translational Molecular Psychiatry Research group, part of the University Hospital of Psychiatry Zurich, is on neurodevelopmental disorders, including attention-deficit hyperactivity disorder (ADHD), Autism spectrum disorders (ASD), psychosis, early onset obsessive-compulsive disorder (OCD) and environmental /stress effects. The lab includes research at the pre-clinical as well as at the basic molecular neuroscience, integrating both research fields in a translational manner. The techniques used include molecular genetics, epigenetic, psychopharmacology, neuronal cellular models and biochemical measures. The goal of the research is to elucidate the etiopathology of the disorders discovering biomarkers for early diagnosis and precision personalised medicine, predicting treatment response and outcomes. Currently, the research group has established patient specific iPSC (induced pluripotent stem cell) neuronal modelling to enable personalized medicine, via studies of the neuronal/molecular alterations in a dish of the disorders using 2D techniques. This will provide a non-invasive approach to investigate etiopathology of neurodevelopmental disorders as well as test drug therapy effects. Methods: Hair follicle Keratinocytes, Prick blood cells culturing, Reprogramming via Sendai, Monolayer Neural Progenitor cells generation, cortical neuronal differentiation Keywords: ADHD, ASD, biochemistry, child and adolescent psychiatry, iPSC, molecular biology, NPC, OCD, psychosis, transcriptomics Topic: Neuroscience, Psychiatry Projects: https://www.kjpd.uzh.ch/de/translationale-molekularpsychiatrie.html Publications: https://www.ncbi.nlm.nih.gov/pubmed/Grünblatt Website: https://www.kjpd.uzh.ch/de/translationale-molekularpsychiatrie.html https://www.ecnp.eu/research-innovation/ECNP-networks/iPSCs-platform ____________________________________________________________________top |
|
|
Prof. Dr. med. Sebastian Jessberger Professor of Neuroscience Brain Research Institute, University of Zurich Research Focus: We use a multipronged, interdisciplinary approach to study the molecular and cellular framework of neural stem cell (NSC) biology in the developing and adult brain. We use state-of-the-art imaging-, genome editing-, and transgenesis-based approaches as well as cellular models of human diseases using pluripotent stem cells with the aim to further our understanding of NSC behavior in health and disease. Methods: Mouse and human embryonic stem cells, genome editing, regionalized forebrain organoids, single cell RNA-sequencing, longitudinal imaging. Keywords: brain development, neurogenesis, asymmetric cell divisions, imaging Topics: Stem cells, neuroscience, brain development, disease modeling Publications: https://pubmed.ncbi.nlm.nih.gov/?term=Jessberger+S&sort=pubdate Website: https://www.hifo.uzh.ch/en/research/jessberger.html ____________________________________________________________________top |
|
|
Prof. Dr. Magdalini Polymenidou Associate Professor of Biomedicine Department of Quantitative Biomedicine, University of Zurich Research Focus: We study the molecular mechanisms of neurodegenerative diseases, focusing on amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). ALS and FTD are characterized by the accumulation of RNA-binding proteins, such as TDP-43 and FUS, as well as non-canonically translated dipeptide repeat proteins (DPRs). The molecular mechanisms that trigger aggregation of ALS/FTD-linked proteins, the basis of disease heterogeneity and the mechanisms of neurotoxicity remain elusive. The aim of our research is to address these important questions using a multidisciplinary approach, working at the molecular, cellular and organismal level. Our vision is to gain deep mechanistic understanding of ALS and FTD to inspire rational design of target-based therapies for these devastating diseases. To explore the disease mechanisms in the context of mature human neurons, we generated highly homogeneous, unmodified, human neural stem cells from iPSCs, along with a robust differentiation method that yields functional, interconnected and long-lived neurons within 3D neural cultures. Inducible expression of ALS/FTD-linked proteins in this system is routinely used to understand their behavior and functional consequences in human neurons, as well as evaluate the therapeutic efficacy of antibodies and other compounds. We are currently expanding our efforts to patient-derived neural systems. Methods: Generation of homogeneous and unmodified human neural stem cells, differentiation to cortical neurons and pure astrocytes, 3D neural cultures, lentiviral generation for inducible expression in neurons, single cell RNA-seq, superresolution microscopy, protein aggregation assays. Keywords: ALS, FTD, TDP-43, FUS, DPRs, LLPS, RNA misregulation, protein aggregation, prion-like Topics: Neuroscience, Neurodegeneration, Brain Diseases, Disease Modelling, RNA biology Publications: https://pubmed.ncbi.nlm.nih.gov/?term=polymenidou%5BAuthor%5D&sort=date Website: https://www.polymenidoulab.com ____________________________________________________________________________ |
|
|
Professor of Systems Neuroscience Institute for Neuroscience, ETH Zurich Research Focus: Despite extensive research, there is up to date no final explanation for the unique cognitive abilities of the human species. Whereas most of the genetic, molecular and cellular components in mammalian brains are quite similar, human neurons seem to display an increased potential for synaptic plasticity. This might be due to a generally higher number of synapses or a prolonged maturation of these in comparison to other mammals and even primates. Non-coding RNAs and especially miRNAs play a significant role in synaptic development and plasticity. Furthermore, particularly the regulatory part of the genome distinguishes humans most from their closest relatives. Nevertheless, the contribution of non-coding RNA dependent mechanisms to human synapse development and plasticity is poorly understood. To study the role of non-coding RNAs in human synapse development, we established a defined human neuronal differentiation protocol without the need for the addition of animal glia cells on two different human induced pluripotent stem cell (iPSC) lines. We show that these neurons are electrophysiological active and investigate synapse formation in detail by analyzing confocal images of pre- and postsynaptic markers at distinct developmental time points. Furthermore, the glia-free differentiation protocol allows us to perform small-RNA sequencing in combination with proteomics and long-RNA sequencing at these same stages. These characterizations are essential for a more detailed functional analysis in the future. By manipulating the expression of specific candidate RNAs, we plan to investigate their role in synaptic maturation as well as effects on neuronal morphology and electrophysiological properties. Newly identified pathways can further be studied in the context of neuropsychiatric diseases using patient-derived iPSC lines. Methods: induced-pluripotent stem cell derived neuron differentiation, imaging, transcriptomics, proteomics, electrophysiology, genome editing Keywords: iPSC, iNeuron, Crispr/Cas9, non-coding RNA, lncRNA, microRNA, synapse, neurodevelopmental disease Topics: Neuroscience, brain development, neuropsychiatric disease Publications: https://pubmed.ncbi.nlm.nih.gov/?term=schratt+g&sort=date Website: http://schrattlab.ethz.ch __________________________________________________________________________top |
|
|
Head of Stem Cell Research Research Focus: We use induced pluripotent stem cells (iPSCs) for modelling and understanding human brain diseases and for regenerative therapies with a focus on Alzheimer’s Disease (AD) and ischemic stroke, respectively. The APOE4 allele is the most important genetic risk factor for Alzheimer’s disease (AD), while the presence of APOE3 is risk-neutral and APOE2 is protective. As the main lipid carrier, APOE has a prominent role in the bioenergetic homeostasis of the brain. However, the role of different APOE isoforms for the metabolic state of human brain cells and for functional crosstalk between neurons and astrocytes is still unknown. Using APOE-isogenic iPSCs, we aim to uncover the underlying mechanism of APOE4-mediated metabolic dysfunctions in differentiated human cortical neurons and astrocytes. Thereby we contribute to the mechanistic understanding of AD pathogenesis and get novel insights into the detrimental role of APOE4 and the protective effects of APOE2. Ischemic troke causes a permanent disability to five million people each year. This is largely due to the lack of effective medical treatments that promote long-term recovery. Therefore, we are developing a next-generation regenerative therapy for stroke based on xeno- and integration-free iPSC-derived neural progenitor cells. These are transplanted into a stroke mouse model and tracked longitudinally using in vivo bioluminescence imaging. Our projects specifically focus on improving applicability, safety and efficacy to advance cell therapy for brain injuries towards potential clinical application. Keywords: Alzheimer’s disease, Stroke, induced pluripotent stem cells (iPSCs), neurodegeneration, neurons, astrocytes, neural progenitor cells, disease modelling, cell transplantation, regenerative medicine. Topics: Neuroscience, Neurodegeneration, Neuroregeneration, Brain Diseases Publications: http://www.ncbi.nlm.nih.gov/pubmed/?term=tackenberg+c Website: www.irem.uzh.ch/Tackenberg.html _________________________________________________________________________top |
|
|
Laboratory of Biosensors and Bioelectronics Institute for Biomedical Engineering ETH Zurich Research Focus: We are promoting a new, bottom-up approach to neuroscience. We create well-defined in vitro neural networks with oriented connections and study their behavior. We use both rat primary hippocampal neurons and human iPSC-derived neurons to build such circuits. We interact with these networks by stimulating selected neurons and record the resulting activities of the networks using optogenetic means and microelectrode arrays (MEAs) including high-density CMOS arrays. We collect TBytes of data that we analyze using machine learning means. We also try to build computational models of the produced living neural networks with the aim to predict the expected behavior of the network. In addition, we also develop stretchable microelectrode arrays to study the strain response of neural networks and the neuro-muscular junction. Methods: Stretchable microelectrode arrays; Optogenetics; Microfabrication; Soft lithography; iPSC-derived neurons Keywords: Neuronal circuits; Induced pluripotent stem cells (iPSCs); Deep learning Topics: Bottom-up Neuroscience; Electrophysiology; Publications: https://lbb.ethz.ch/publications/papers.html Website: https://lbb.ethz.ch/ _______________________________________________________________________top |







